|
|
GOEAST -- Gene Ontology Enrichment Analysis Software Toolkit
GOEAST is web based software toolkit providing easy to use, visualizable, comprehensive and unbiased Gene Ontology (GO) analysis for high-throughput experimental results, especially for results from microarray hybridization experiments. The main function of GOEAST is to identify significantly enriched GO terms among give lists of genes using accurate statistical methods.
Read more about how to use GOEAST...
Curent GOEAST server loading factor: 0 (idle)
Advantages and Unique Features
Compared with available GO analysis tools, GOEAST has the following unique features:
-
GOEAST supports analysis for data from various resources, such as expression data obtained using Affymetrix, illumina, Agilent or customized microarray platforms. GOEAST also supports non-microarray based experimental data. The web-based feature makes GOEAST very user friendly; users only have to provide a list of genes in correct formats.
-
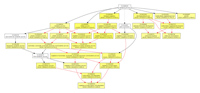
GOEAST provides visualizable analysis results, by generating graphs exhibiting enriched GO terms as well as their relationships in the whole GO hierarchy. See
Fig 1 as an example.
Fig 1. Example of GOEAST graphical results
|
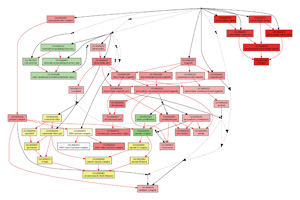
Fig 2. Example of Multi-GOEAST graphical results
|
Note that GOEAST generates separate graph for each of the three GO categories, namely
biological process, molecular function and cellular component.
-
GOEAST allows comparison of results from multiple experiments (see
Multi-GOEAST tool). The displayed color of each GO term node in graphs generated by Multi-GOEAST is the combination of different colors used in individual GOEAST analysis. See
Fig 2 as an example.
Try GOEAST now!
GOEAST Latest News
- GOEAST now shows the level for each GO terms! The level is defined in
FAQ.
- Advanced Paramter Settings of all GOEAST now allows more p-value/FDR cut-offs!
- GOEAST is even faster now! We have upgraded both the algorithm and the underlying database to make GOEAST now about 20 times faster than before!
- GOEAST Agilent® tool now supports Agilent Array Names as well as the Array-IDs! You can find you Agilent array platform much easier! Try it now!
- GOEAST has upgraded to a much powerful server now!
- Graphic size and witdh:height ratio is better controlled now for all GOEAST tools!
- GOEAST supports more advanced graphic parameters in the advanced settings now!
Try it now!
- Now you can provide an optional annotation field for your customized probes in
GOEAST customized microarray analysis tool for convenience.
Try it now!
- Few microarray platforms contain probes / probe-sets for more than one species, and GOEAST Affymetrix tool supports both per-chip backgrounds and per-species
backgrounds now! Try it now!
- GOEAST supports official gene symbols in Batch-Genes tool now!
Try it now!
Tip of using GOEAST
Choose JPEG over PDF format of output figures, whenever you cannot see your PDF result figures.
Choose a non-busy time when using GOEAST. Check the loading factor first at the homepage before starting your jobs.
Try to control the graphic size by setting detail information level, font-size and
graphic-condensing method in the advanced parameter settings.
Choose your favourite statistical test method, multi-test adjustment approach and significance
level in the advanced parameter settings.
Try GOEAST Customized Microarray Analysis if you could not find support of your species or you want to analyze genes agains customized reference genes.
Name your job before start each GOEAST analysis! Multiple GOEAST analysis results will not
be messed up any more.
Reference
Please cite the following article if you found GOEAST helpful in your work:
Nucleic Acids Res. 2008 May 16.
GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis.
Zheng Q, Wang XJ.
PMID: 18487275
|
GOEAST News:
2016-07-02: new!
GOEAST server auto update is significantly speeded up, i.e. much shorter maintenance time now.
2018-09-17:
Gene Ontology information updated.
2018-09-10:
Gene Ontology information updated.
2018-09-03:
Gene Ontology information updated.
2018-08-27:
Gene Ontology information updated.
2018-08-20:
Gene Ontology information updated.
2018-08-13:
Gene Ontology information updated.
2018-08-06:
Gene Ontology information updated.
2018-07-30:
Gene Ontology information updated.
2018-07-23:
Gene Ontology information updated.
2018-07-16:
Gene Ontology information updated.
2018-07-09:
Gene Ontology information updated.
2018-07-02:
Gene Ontology information updated.
2018-06-25:
Gene Ontology information updated.
2018-06-18:
Gene Ontology information updated.
2018-06-11:
Gene Ontology information updated.
More news for GOEAST ...
|
 Home
Home Tools
Tools Statistics
Statistics Advance
Advance Tutorial
Tutorial FAQ
FAQ Contact us
Contact us Home
Home Tools
Tools Statistics
Statistics Advance
Advance Tutorial
Tutorial FAQ
FAQ Contact us
Contact us

