Tutorial
GOEAST is a very user-friendly toolkit. You can finish your analysis in a few simple steps.
Use GOEAST tools
The GOEAST tools web page provides tools for analyzing microarray experiment results from various array platforms, including both commercial and customized microarrays. To lauch the "GOEAST tools" web page, click the
"Tools" button (marked area in Fig 1) at the top of GOEAST web pages.
Fig 1. Header of GOEAST pages

|
At the "GOEAST tools" page, you could find the corresponding tool for the various microarray platforms. Currently, GOEAST has tools for three commercial microarray companies (
Affymetrix,
illumina and
Agilent) and for any customized microarray,
if probes on the customized microarray have GO annotation information. For example, if you used the
Affymetrix® microarrays, you could find tool for them at marked area in Fig 2.
Fig 2. GOEAST tools page

|
After choosing the right microarray platform, you could easily start analysis following instructions
provided on each page. For example, if you are at the page of GOEAST Affymetrix tool,
you have to select the species and the microarray name you used first, then provide the gene list in
Affymetrix probeset ID format either by cut-and-paste method or by uploading a plain-text file.
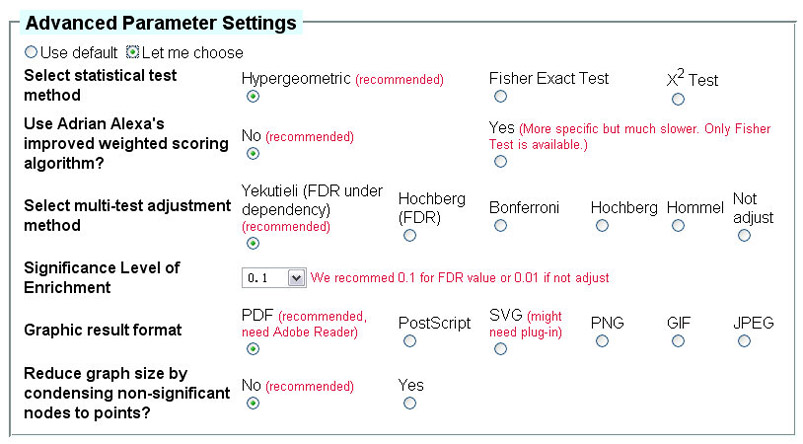
Parameters related to analysis results could be changed after selecting the "Let me choose" option
(Fig 3). Since some analysis might take up to an hour, a valid email address
is required to send you the results.
Fig 3. Advanced parameters available for GOEAST tools
|
The results of GOEAST analysis will be displayed in the processing page
(Fig 4). An email contains link to the results will also be sent to users
(Fig 5).
Fig 4. Example of waiting pages for GOEAST tools

|
Fig 5. Example of finishing email for GOEAST tools

|
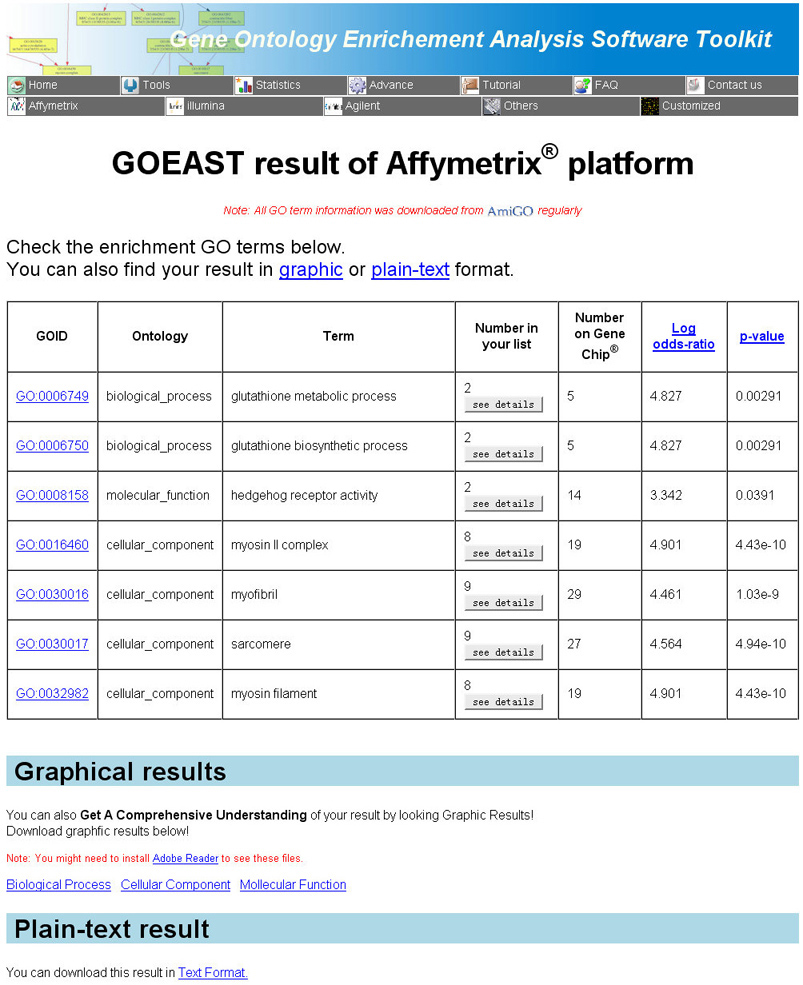
Ok, now we come to the GOEAST result part. GOEAST result pages
provides outputs in three different formats: a graphical file for each GO category, a HTML table summarizing
all enriched GO terms, and a plain-text file (Fig 6).
All GOEAST results could be downloaded, the plain-text file will be needed if you want to do Multi-GOEAST analysis as well. To kown more about the GOEAST results,
click here.
Fig 6. Example of GOEAST analysis results
|
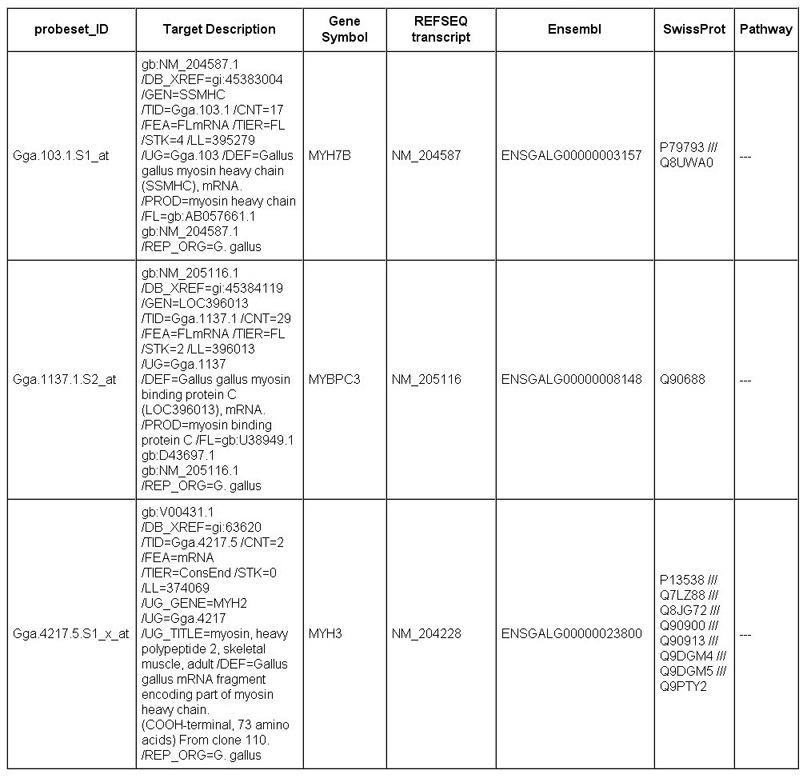
The "see details" button in the HTML table provides additional information of each enriched GO terms.
The content of information provided by this function might vary among different microarray platforms,
according to the availability of information associated with each platform and the space limitation.
For example, detailed information of the Affymetrix microarray analysis results contains the probeset IDs,
probeset target description, gene symbol, REFSEQ transcript accession, Ensembl ID,
SwissProt ID and KEGG pathway information. (Fig 7).
Fig 7. Example of a enriched GO term detail of Affymetrix platform
|
Use GOEAST Advanced tools
Currently, two advanced tools are available, Batch-Genes and Multi-GOEAST. The Batch-Genes is an advanced tool providing similar GO enrichment analysis for
non-array based experimental results. NCBI REFSEQ IDs are required for Batch-Genes
analysis.
See more description of Batch-Genes tool.
The Muiti-GOEAST is an advanced tool for comparison of several GOEAST
analysis results.
See more description of Multi-GOEAST tool.
To use GOEAST advanced tools, click the "Advance" button at the top of
GOEAST pages (Fig 1).
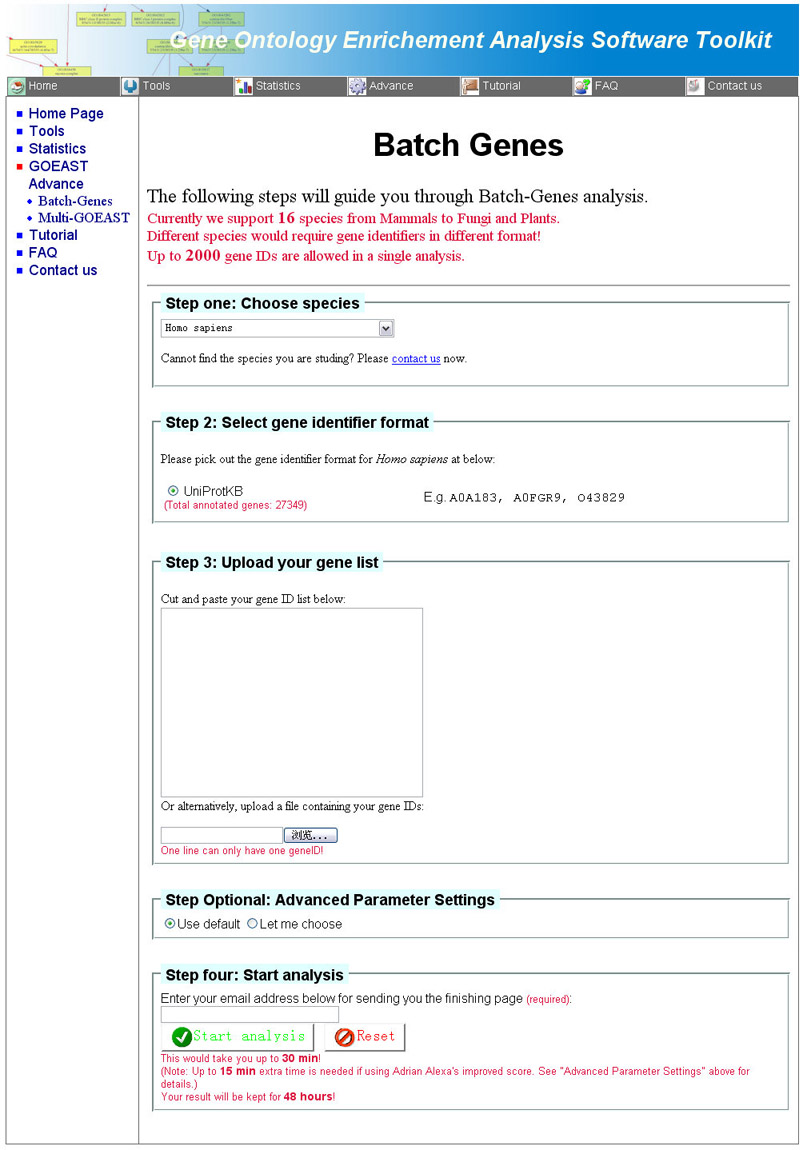
The process of using Batch-Genes analysis is similar to that of GOEAST
analysis. You only need to select the correct species, upload your gene list in NCBI REFSEQ transcript accession format, and provide a valid email address for recieving results.
(Fig 8). The advanced parameter settings are identical to GOEAST
tools for microarray analysis (Fig 3).
Fig 8. The Batch-Genes tool page
|
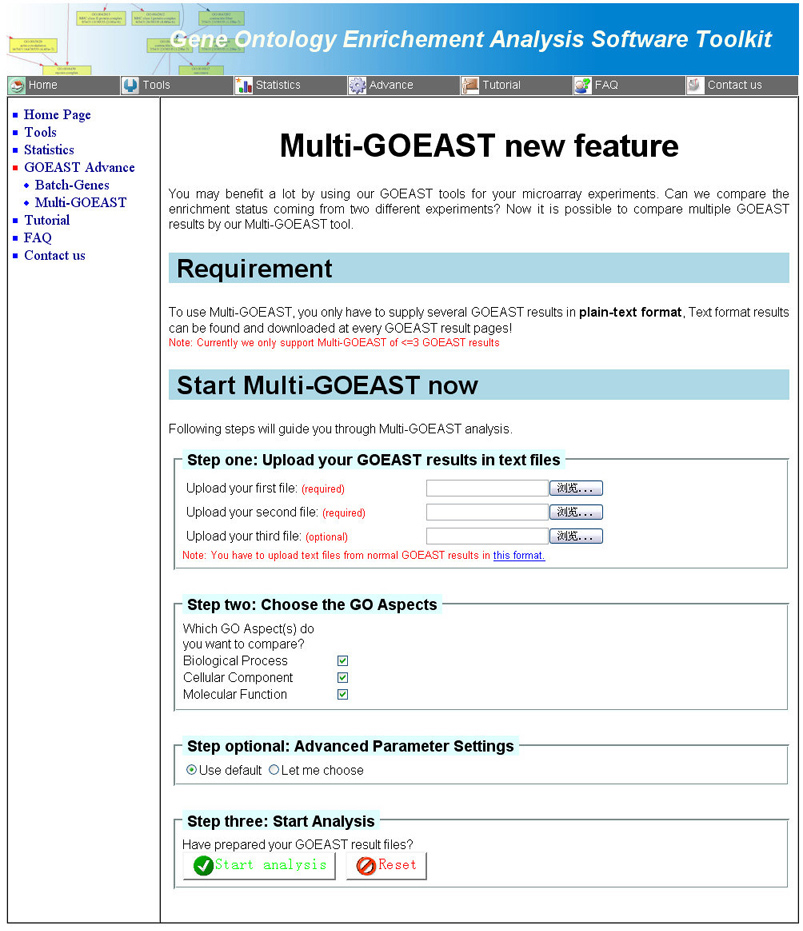
The Multi-GOEAST tool compares GO term enrichment status of different
GOEAST outputs, therefore, the plain-text output file of GOEAST tools
is required to use Multi-GOEAST function (Fig 9).
All three GO categories would be compared by default. To accelerate the analysis process, users can also
define certain GO categories to compare.
Fig 9. The Multi-GOEAST tool page
|
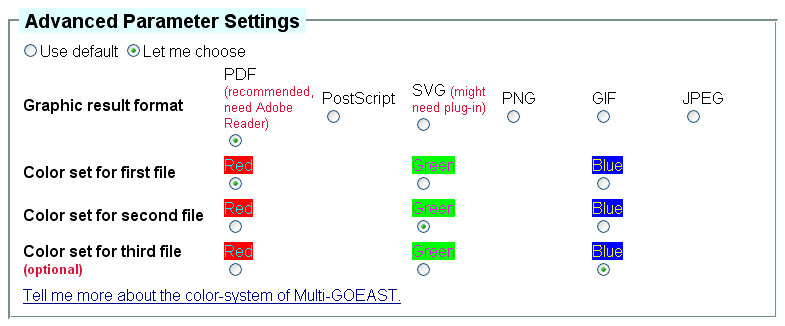
Advanced parameter settings for Multi-GOEAST tool is also different from other
GOEAST tools. You can select your favorite graphic format as well the color combinations
to generate the graphic outputs (Fig 10). Since Multi-GOEAST
usually finishes in a short time, the results will be displayed on the web page directly, no email address
is required.
Fig 10. Advanced parameters available for Multi-GOEAST tool.
|
Please see FAQs for explainations of
GOEAST results.
|
 Home
Home Tools
Tools Statistics
Statistics Advance
Advance Tutorial
Tutorial FAQ
FAQ Contact us
Contact us Home
Home Tools
Tools Statistics
Statistics Advance
Advance Tutorial
Tutorial FAQ
FAQ Contact us
Contact us