Manual: Stem-loop Small RNA Prediction
The "Stem-loop Small RNA Prediction" function can be launched by clicking corresponding button at the navigation bar of psRobot web page (Fig. 1).

Fig. 1. Click the tab in orange box to launch the stem-loop small RNA prediction function.
On the module page (Fig. 2), you need to upload one or several small RNA sequencs in FASTA format or plain text format (Fig. 3), and choose a correct species from which your small RNA sequences are obtained. The demo small RNAs and the demo selection of genome will be presented by clicking the demo buttons above the widgets.
Check the checkbox with the label "Show small RNA conservation information for each submitted sequence in the result" (underlined in Fig. 2) will make the server do the small RNA conservation analysis in 8 plant species as shown in the help index page. But the selection will slow down the processing (about 30 seconds for one small RNA sequence), thus is unchecked in default. You can check it anyway if you want the see the information.
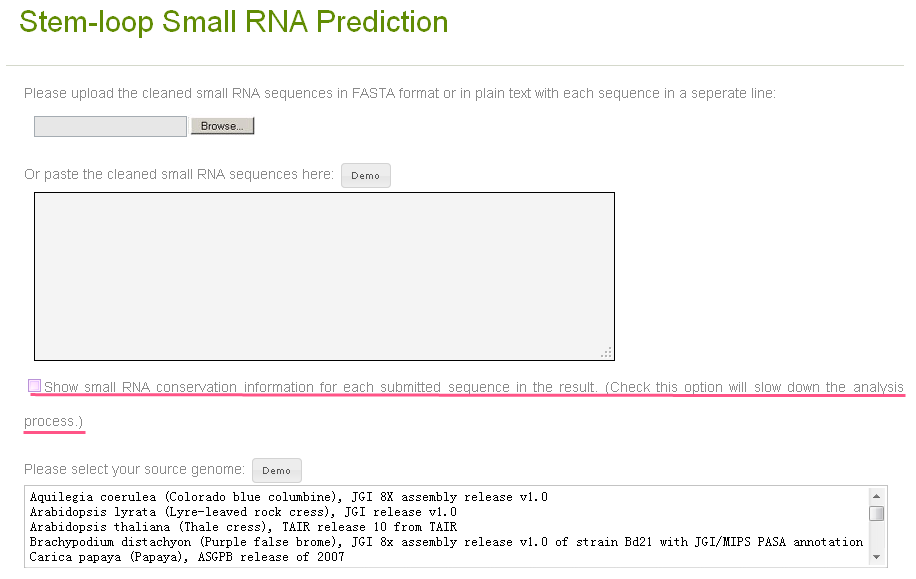
Fig. 2. Stem-loop small RNA prediction tool page.

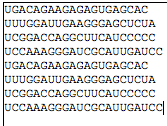
Fig. 3. The sample input small RNAs. The server allows input either in FASTA format (left) or in plain text with each sequence in a seperated line (right).
We designed 4 parameters to make the stringency of the stem-loop small RNA prediction tunable. There are two sets of default parameters that can be selected by clicking the "Strict" and "Loose" buttons above the parameter form (Fig. 4). The strict option allows larger range of mismatch number and discards long loop miRNAs (Fig. 5), while the loose option vise versa. The adjustable parameters for the 'stem-loop small RNA prediction' function (Fig. 4) are:
- Minimal number of mismatches in small RNA region: minimal number of mismatched nucleotides allowed in the query small RNA region. The strict option sets it as '1' and the loose one sets it as '0'.
- Maximal number of mismatches in small RNA region: maximal number of mismatched nucleotides allowed in the query small RNA region. Both options set it as '7'.
- Retain long loop small RNAs [T/F]: Majority of the canonical miRNAs have a short loop region in their stem-loop shaped precursor structures, as examplified in Fig. 5(a), but there are some miRNAs and other stem-loop small RNAs with a long loop region in their stem-loop shaped precursor structures, as examplified in Fig. 5(b). Choose 'T' or 'F' to decide whether to allow long loop precursors or not. The strict default is 'F' and the loose is "T".
- Maximal precursor length: The maximal length of extracted small RNA surrounding genomic sequences used in stem-loop precursor structure prediction. Larger value will take longer running time, but retain longer precursors in the results. The strict default is '100' and the loose default is '200'.

Fig. 4. Parameter setting options of the stem-loop small RNA prediction tool.
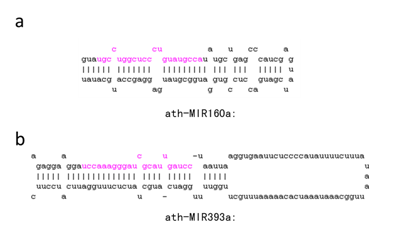
Fig. 5. Examples of predicted precursor structures of known plant miRNAs with short (a) and long (b) loop regions.
The result of the small RNA stem-loop prediction function includes:
- The expression abundance of query small RNA in different small RNA biogenesis related gene mutants and AGO protein complexes (Fig. 6(1)). Numbers shown in the result table are sequence reads in each datasets after normalized to RPM (Reads Per Million reads). Information of all included datasets is detailed in Appendix I.
- The conservation status of the query small RNA in 8 plant species (4 monocots and 4 dicots): Arabidopsis thaliana (Ath), Brachypodium distachyo(Bdi), Carica papaya (Cpa), Oryza sativa (Osa), Populus trichocarpa (Ptr), Sorghum bicolor (Sbi), Vitis vinifera (Vvi), and Zea mays (Zma). And a multiple sequence alignment of the query small RNA and its ortholog sequences in conserved species. In the table, the query small RNA is conserved in the species with a "+" symbol in their columns, and not conserved in the "-" columns. "qs" indicates the species the query small RNA sequence is from (Fig. 6(2)).
- The number of perfectly matched genomic locations of the query sequences, the detailed information of each loci, and the number of predicted stem-loop shaped precursors (Fig. 6(3)). If the small RNA is from a repetitive genomic region, the repeat name and type of that region will be shown in the rightmost column of the table.
- The detailed information of each genomic location of the query stem-loop small RNAs; the extracted small RNA precursors; the predicted precursor secondary structures and their folding energy (Fig. 6(4)). Click the link of "Predict Targets for The Small RNA" will redirect to the target prediction page with the small RNA sequence set as the query small RNA.
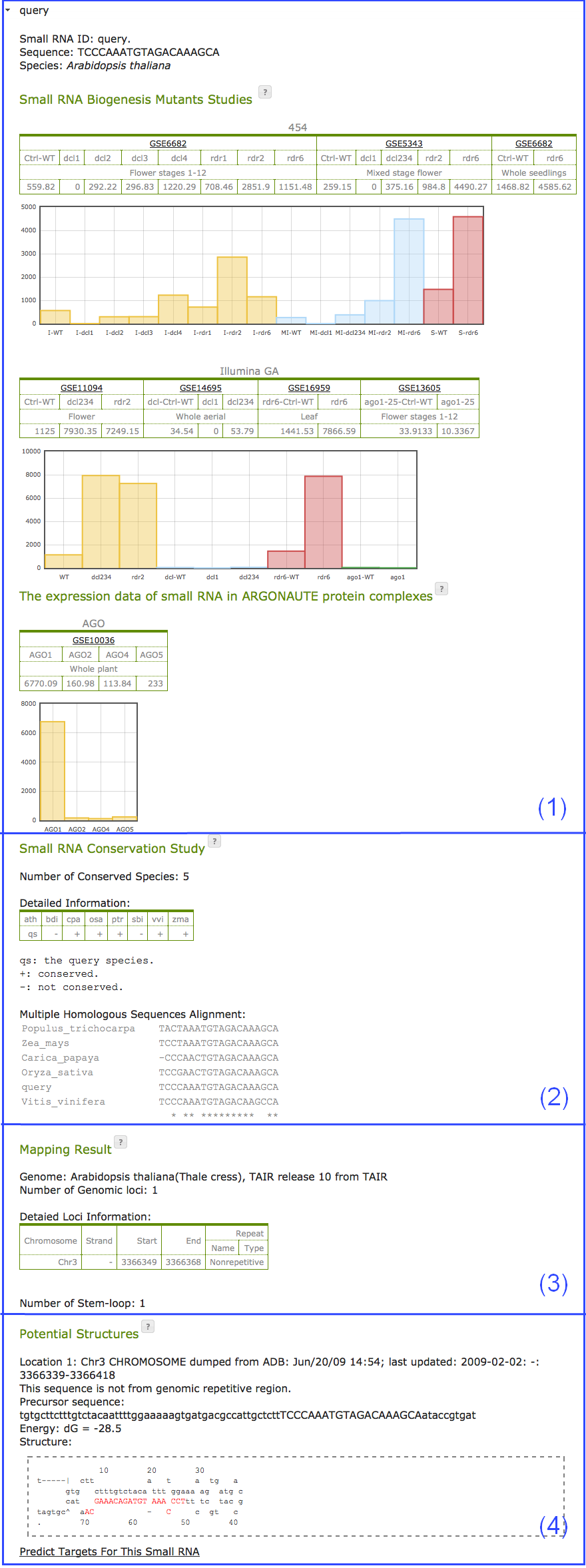
Fig. 6. Result page of the small RNA stem-loop prediction function. Click here for an interactive sample output.
Appendix I. Collected small RNA deep-sequencing datasets from small RNA biogenesis-related gene mutants and AGO protein complexes:
Seven datasets containing small RNA data from small RNA biogenesis related gene mutants and AGO protein complexes are used for analysis of Arabidopsis thaliana small RNAs.
| Sample name | Sample type | Tissue | GEO dataset |
| 454-I-Ctrl-WT | WT | Flower stages 1-12 | GSE6682 |
| 454-I-dcl1-7 | dcl1-7 | Flower stages 1-12 | GSE6682 |
| 454-I-dcl2-1 | dcl2-1 | Flower stages 1-12 | GSE6682 |
| 454-I-dcl3-1 | dcl3-1 | Flower stages 1-12 | GSE6682 |
| 454-I-dcl4-2 | dcl4-2 | Flower stages 1-12 | GSE6682 |
| 454-I-rdr1-1 | rdr1-1 | Flower stages 1-12 | GSE6682 |
| 454-I-rdr2-1 | rdr2-1 | Flower stages 1-12 | GSE6682 |
| 454-I-rdr6-15 | rdr6-15 | Flower stages 1-12 | GSE6682 |
| 454-MI-Ctrl-WT | WT | Mixed stage flower | GSE5343 |
| 454-MI-dcl1-7 | dcl1-7 | Mixed stage flower | GSE5343 |
| 454-MI-dcl234 | dcl234 | Mixed stage flower | GSE5343 |
| 454-MI-rdr2 | rdr2 | Mixed stage flower | GSE5343 |
| 454-MI-rdr6 | rdr6 | Mixed stage flower | GSE5343 |
| 454-S-Ctrl-WT | WT | whole seedlings | GSE6682 |
| 454-S-rdr6-15 | rdr6-15 | whole seedlings | GSE6682 |
| SBS-I-Ctrl-WT | WT | Flower | GSE11094 |
| SBS-I-dcl234 | dcl234 | Flower | GSE11094 |
| SBS-I-rdr2 | rdr2 | Flower | GSE11094 |
| dcl-Ctrl-WT | WT | Whole aerial | GSE14695 |
| dcl1 | dcl1-7 | Whole aerial | GSE14695 |
| dcl234 | dcl2-1, dcl3-1, dcl4-2 | Whole aerial | GSE14695 |
| rdr6-Ctrl-WT | WT | Leaf | GSE16959 |
| rdr6 | rdr6-15 | Leaf | GSE16959 |
| ago1-25-Ctrl-WT | WT | Flower stages 1-12 | GSE13605 |
| ago1-25 | ago1-25 | Flower stages 1-12 | GSE13605 |
| AGO1 | AGO1 associated sRNAs | Whole plant | GSE10036 |
| AGO2 | AGO2 associated sRNAs | Whole plant | GSE10036 |
| AGO4 | AGO4 associated sRNAs | Whole plant | GSE10036 |
| AGO5 | AGO5 associated sRNAs | Whole plant | GSE10036 |
Two datasets (GSE20748 and GSE18250) containing dcl1, dcl3, rdr2 mutants and AGO1a/b/c, AGO4a/b and AGO16 associated small RNA data are used for analysis of Oryza sativa small RNAs.
| Sample name | Sample type | Tissue | GEO dataset |
| WT1 | WT | Whole plant | GSE20748 |
| dcl1 | dcl1 | Whole plant | GSE20748 |
| dcl3 | dcl3 | Whole plant | GSE20748 |
| rdr2 | rdr2 | Whole plant | GSE20748 |
| AGO4a | AGO4a associated sRNAs | Whole plant | GSE20748 |
| AGO4b | AGO4b associated sRNAs | Whole plant | GSE20748 |
| AGO16 | AGO16 associated sRNAs | Whole plant | GSE20748 |
| WT2 | Total sRNAs | Whole plant | GSE18250 |
| AGO1a | AGO1a associated sRNAs | Whole plant | GSE18250 |
| AGO1b | AGO1b associated sRNAs | Whole plant | GSE18250 |
| AGO1c | AGO1c associated sRNAs | Whole plant | GSE18250 |